Simulates canopy assimilation for C3 canopies
Usage
c3CanA(lai, doy, hr, solar, temp, rh, windspeed, lat = 40, nlayers = 8, kd = 0.1, heightFactor = 3, c3photoControl = list(), lnControl = list(), StomWS = 1)
Arguments
- lai
- leaf area index.
- doy
- day of the year, (1--365).
- hr
- hour of the day, (0--23).
- solar
- solar radiation (
micromolm-2s-1). - temp
- temperature (Celsius).
- rh
- relative humidity (0--1).
- windspeed
- wind speed (m
s-1). - lat
- latitude.
- nlayers
- number of layers in the simulation of the canopy (max allowed is 50).
- kd
- Ligth extinction coefficient for diffuse light.
- heightFactor
- Height Factor. Divide LAI by this number to get the height of a crop.
- c3photoControl
- list that sets the photosynthesis parameters for c3 plants through the c3photoParms function
- lnControl
- list that sets the leaf nitrogen parameters. LeafN: Initial value of leaf nitrogen (g m-2). kpLN: coefficient of decrease in leaf nitrogen during the growing season. The equation is LN = iLeafN * exp(-kLN * TTc). lnFun: controls whether there is a decline in leaf nitrogen with the depth of the canopy. 'none' means no decline, 'linear' means a linear decline controlled by the following two parameters. lnb0: Intercept of the linear decline of leaf nitrogen in the depth of the canopy. lnb1: Slope of the linear decline of leaf nitrogen in the depth of the canopy. The equation is 'vmax = leafN_lay * lnb1 + lnb0'.
Value
list
returns a list with several elements
CanopyAssim: hourly canopy assimilation (Mg/ha per hour)
CanopyTrans: hourly canopy transpiration (Mg/ha per hour)
CanopyCond: hourly canopy conductance (units ?)
TranEpen: hourly canopy transpiration according to Penman
(Mg ha^{-1} per hour)
TranEpen: hourly canopy transpiration according to Priestly
(Mg/ha per hour)
LayMat: hourly by Layer matrix containing details of the calculations by layer (each layer is a row). col1: Direct Irradiance col2: Diffuse Irradiance col3: Leaf area in the sun col4: Leaf area in the shade col5: Transpiration of leaf area in the sun col6: Transpiration of leaf area in the shade col7: Assimilation of leaf area in the sun col8: Assimilation of leaf area in the shade col9: Difference in temperature between the leaf and the air (i.e. TLeaf - TAir) for leaves in sun. col10: Difference in temperature between the leaf and the air (i.e. TLeaf - TAir) for leaves in shade. col11: Stomatal conductance for leaves in the sun col12: Stomatal conductance for leaves in the shade col13: Nitrogen concentration in the leaf (g m^-2) col14: Vmax value as depending on leaf nitrogen
Description
It represents an integration of the photosynthesis function
c3photo, canopy evapo/transpiration and the
multilayer canopy model sunML.
Details
The photosynthesis function is modeled after the version in WIMOVAC. This is based on the well known Farquar model.
Note
References
Farquhar model site here ~
Examples
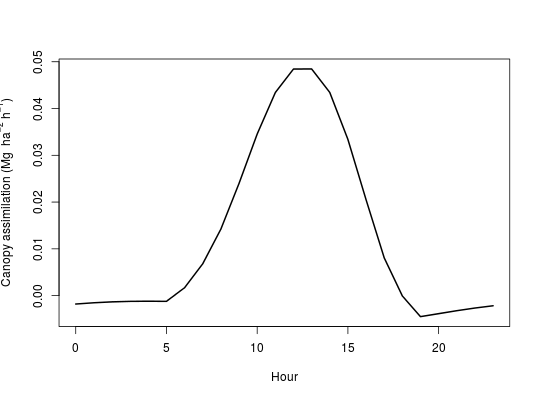
data(doy124) tmp <- numeric(24) for(i in 1:24){ lai <- doy124[i,1] doy <- doy124[i,3] hr <- doy124[i,4] solar <- doy124[i,5] temp <- doy124[i,6] rh <- doy124[i,7] ws <- doy124[i,8] tmp[i] <- c3CanA(lai,doy,hr,solar,temp,rh,ws)$CanopyAssim } plot(c(0:23),tmp, type='l',lwd=2, xlab='Hour', ylab=expression(paste('Canopy assimilation (Mg ', ha^-2,' ',h^-1,')')))